CRSU Apps
MetaInsight Covid-19
Network Meta-Analysis of Pharmacological treatments for COVID 19: Tool for exploration, re-analysis, sensitivity analysis, and interrogation of data from living systematic reviews
https://crsu.shinyapps.io/metainsightcovid/
MetaInsight
https://crsu.shinyapps.io/metainsight/
MetaInsight is a new tool that is freely available and that conducts network meta-analysis (NMA) via the web requiring no specialist software for the user to install but leveraging established analysis routines (specifically the netmeta and gemtc package in R). The tool is interactive and uses an intuitive ‘point and click’ interface and presents results in visually intuitive and appealing ways. It can also carry out sensitivity analysis on existing NMAs. It is hoped that this tool will assist those in conducting NMA who are not statistical experts, and, in turn, increase the relevance of published meta-analyses, and in the long term contribute to improved healthcare decision making as a result.
If you use the app please cite it as: Owen, RK, Bradbury, N, Xin, Y, Cooper, N, Sutton, A. MetaInsight: An interactive web‐based tool for analyzing, interrogating, and visualizing network meta‐analyses using R‐shiny and netmeta. Res Syn Meth. 2019; 10: 569– 581. https://doi.org/10.1002/jrsm.1373
New Treatment Ranking Feature
https://crsu.shinyapps.io/MetaInsight_Beta/
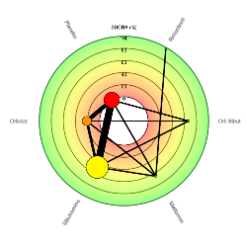
The ability to rank treatments within a network is a hugely advantageous benefit of conducting NMAs, however the visualisation of ranking results can often lead to misleading conclusions. To this end, the CRSU set out to redesign the ranking features within MetaInsight.
The Beta version of MetaInsight as linked above contains an entire 'ranking panel' where Bayesian analyses are run. This panel contains the ranking results alongside relative effects and network summaries to aid interpretation and encourage sensible conclusions. Furthermore, the panel includes two newly developed plots to present the ranking results themselves, including options such as colour-blind friendly versions.
If you have any questions or feedback regarding this new feature, please do email Clareece Nevill at clareece.nevill@le.ac.uk
MetaInsight V1.1 (Frequentist analysis)
Binary : https://crsu.shinyapps.io/metainsightb/
Continuous: https://crsu.shinyapps.io/metainsightc/
DTA Apps
DTA-MA: https://crsu.shinyapps.io/dta_ma/
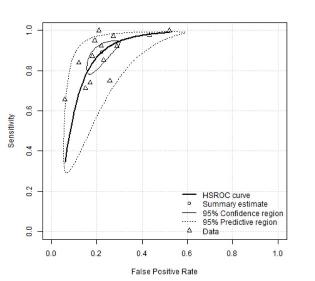
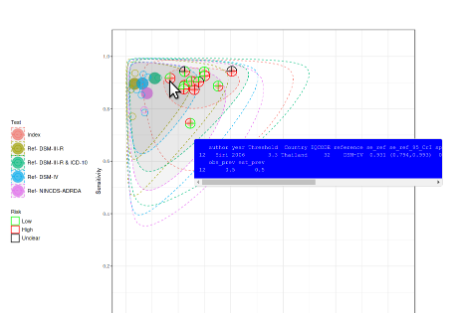
This app provides a user friendly (“point and click”) web interface for conducting meta-analysis of diagnostic test accuracy (DTA) studies. It uses macros written (by others) in R “behind the scenes” to achieve this. Analysis options include the bivariate meta-analysis model - recommended for use in Cochrane DTA reviews (but not available in Cochrane software). It can also be used to carry out sensitivity analysis on existing meta-analyses. All results produced are downloadable, including high quality graphical plots. It is hoped that this tool will assist those in conducting NMA who are not statistical experts, and, in turn, increase the relevance of published meta-analyses, and in the long term contribute to improved healthcare decision making as a result.
If you use this app please cite it as:
Freeman SC, Kerby CR, Patel A, Cooper NJ, Quinn T, Sutton AJ. Development of an interactive web-based tool to conduct and interrogate meta-analysis of diagnostic test accuracy studies: MetaDTA. BMC Medical Research Methodology 2019; 19: 81 https://doi.org/10.1186/s12874-019-0724-x
DTA Primer: https://crsu.shinyapps.io/diagprimer/
This interactive explorable explanation is designed to teach the basics of diagnostic test accuracy evaluation and is recommended for anyone new to the area (including those planning to move into diagnostic test synthesis) or those who never quite got their head around an ROC curve or needs a bit of revision on the topic.
A smoother implementation of the core idea in the above is now available here (Note: You will probably still want to look at the initial app for the explanations and extra content etc)
MetaBayesDTA (BETA v1.0)
https://crsu.shinyapps.io/MetaBayesDTA/
MetaBayesDTA is an extended, Bayesian version of MetaDTA (https://crsu.shinyapps.io/dta_ma/), which allows users to conduct meta-analysis of test accuracy, with or without assuming a gold standard. Due to its user-friendliness and broad array of features, MetaBayesDTA should appeal to a wide variety of applied researchers, including those who do not have the specific expertise required to fit such models using statistical software. Furthermore, MetaBayesDTA has many features not available in other apps. For instance, for the bivariate model, one can conduct subgroup analysis and univariate meta-regression. Meanwhile, for the model which does not assume a perfect gold standard, the app can partially account for the fact that different studies in a meta-analysis often use different reference tests using meta-regression.
Please note that the current release is a Beta version and does not yet have a manual. Please send questions and any feedback - including bug reports and suggestions for new features - to Enzo Cerullo (enzo.cerullo@bath.edu).
We kindly ask you to cite this app as:
Cerullo E, Freeman SC, Kerby CR, Sutton AJ, Cooper NJ, Quinn TJ, Wu O. MetaBayesDTA: Bayesian meta-analysis of diagnostic test accuracy data, with or without a gold standard. NIHR Complex Reviews Support Unit (CRSU).
Furthermore, please cite the following papers associated with the previous version of the app - MetaDTA - whenever outputs from MetaBayesDTA are used:
Patel A, Cooper NJ, Freeman SC, Sutton AJ. Graphical enhancements to summary receiver operating charcateristic plots to facilitate the analysis and reporting of meta-analysis of diagnostic test accuracy data. Research Synthesis Methods 2020, https://doi.org/10.1002/jrsm.1439.
Freeman SC, Kerby CR, Patel A, Cooper NJ, Quinn T, Sutton AJ. Development of an interactive web-based tool to conduct and interrogate meta-analysis of diagnostic test accuracy studies: MetaDTA. BMC Medical Research Methodology 2019; 19: 81